Sbiojn--ReactionNetwork
Python Package. December 13, 2017
By Joely Nelson and Eric Klavins
A python framework that uses numpy and sypy to make it easy to quickly generate and analyze chemical reaction networks. This package was created by expanding off of Eric Klavins’s code for CSE 486.
It allows the user to model chemical reaction networks, automatically generate stoichiometric matrices, analyze the equilibrium, graph uncertainty, and perform other mathematical operations
With just a few lines of code a user can create a model of the central dogma of biology: RNA being generated, and that RNA becoming protein. Below is an example of this model. The x axis represents time, the y axis represents the amount, and the shaded areas represent the uncertainty and noise of what is predicted.
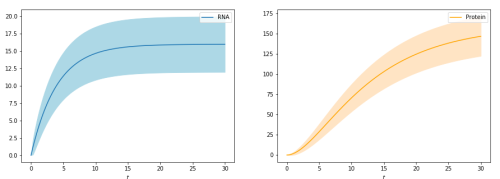
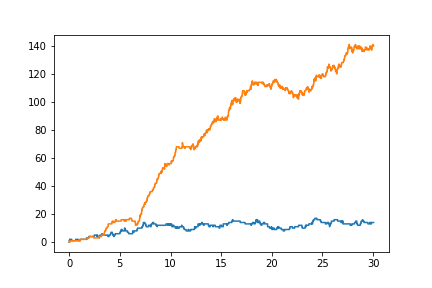
The user below has used Sbjiojn to model the same system above, but has used random state jumping to observe the system uncertainty.
You can go to https://gitlab.cs.washington.edu/joelyn/sbiojn to download the framework for yourself.